Clipping data with polygons
This exercise uses PDAL to apply to clip data with polygon geometries.
Note
This exercise is an adaption of the PDAL tutorial.
Exercise
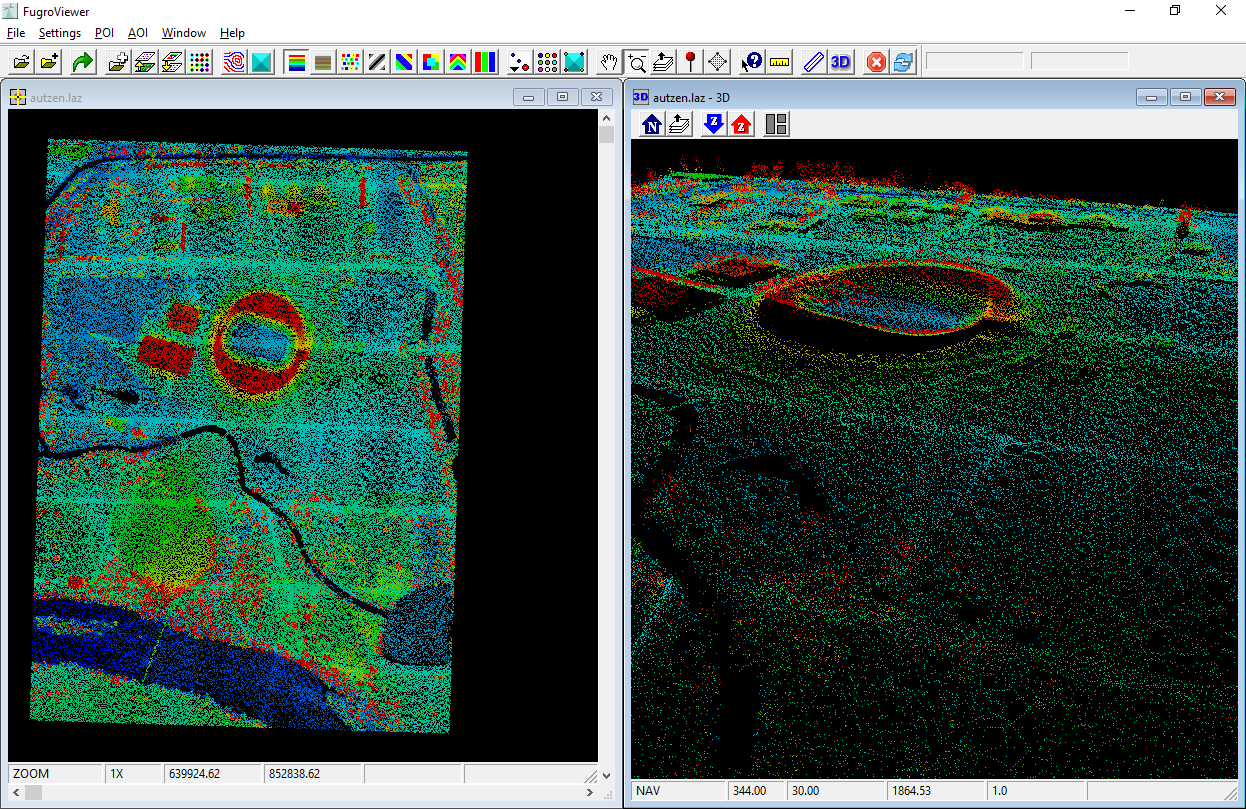
The autzen.laz file is a staple in PDAL and libLAS examples. You can
download this file here. We will use
this file to demonstrate clipping points with a geometry. We’re going to clip
out the stadium into a new LAS file.
Data preparation
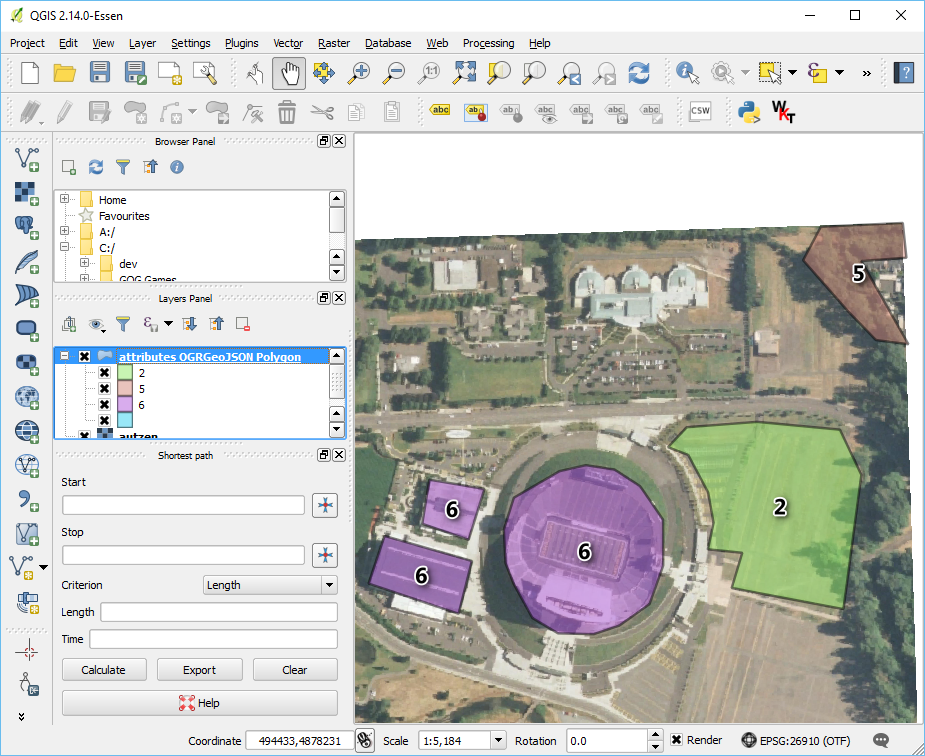
The data are mixed in two different coordinate systems. The LAZ file is in Oregon State Plane Ft. and the GeoJSON defining
the polygons, attributes.json, is in EPSG:4326. We have two options –
project the point cloud into the coordinate system of the attribute polygons,
or project the attribute polygons into the coordinate system of the points. The
latter is preferable in this case because it will be less math and therefore
less computation. To make it convenient, we can utilize OGR’s VRT
capability to reproject the data for us on-the-fly:
<OGRVRTDataSource>
<OGRVRTWarpedLayer>
<OGRVRTLayer name="OGRGeoJSON">
<SrcDataSource>./exercises/analysis/clipping/attributes.json</SrcDataSource>
<SrcLayer>attributes</SrcLayer>
<LayerSRS>EPSG:4326</LayerSRS>
</OGRVRTLayer>
<TargetSRS>+proj=lcc +lat_1=43 +lat_2=45.5 +lat_0=41.75 +lon_0=-120.5 +x_0=399999.9999999999 +y_0=0 +ellps=GRS80 +units=ft +no_defs</TargetSRS>
</OGRVRTWarpedLayer>
</OGRVRTDataSource>
Note
This VRT file is available in your workshop materials in the
./exercises/analysis/clipping/attributes.vrt file. You will need to
open this file, go to line 4 and replace ./ with
the correct path for your machine.
A GDAL or OGR VRT is a kind of “virtual” data source definition type that combines a definition of data and a processing operation into a single, readable data stream.
Note
The GeoJSON file does not have an externally-defined coordinate system, so we are explicitly setting one with the LayerSRS parameter. If your data does have coordinate system information, you don’t need to do that. See the OGR VRT documentation for more details.
Pipeline breakdown
{
"pipeline": [
"./exercises/analysis/clipping/autzen.laz",
{
"column": "CLS",
"datasource": "./exercises/analysis/clipping/attributes.vrt",
"dimension": "Classification",
"layer": "OGRGeoJSON",
"type": "filters.overlay"
},
{
"limits": "Classification[6:6]",
"type": "filters.range"
},
"./exercises/analysis/clipping/stadium.las"
]
}
Note
This pipeline is available in your workshop materials in the
./exercises/analysis/clipping/clipping.json file. Remember
to replace each of the three occurrences of ./
in this file with the correct location for your machine.
1. Reader
autzen.laz is the LASzip file we will clip.
2. filters.overlay
The filters.overlay filter allows you to assign values for coincident
polygons. Using the VRT we defined in Data preparation,
filters.overlay will
assign the values from the CLS column to the Classification field.
3. filters.range
The attributes in the attributes.json file include polygons with values
2, 5, and 6. We will use filters.range to keep points with
Classification values in the range of 6:6.
4. Writer
We will write our content back out using a writers.las.
Execution
Invoke the following command, substituting accordingly, in your Conda Shell:
The –nostream option disables stream mode. The point-in-polygon check (see notes) performs poorly in stream mode currently.
$ pdal pipeline ./exercises/analysis/clipping/clipping.json --nostream
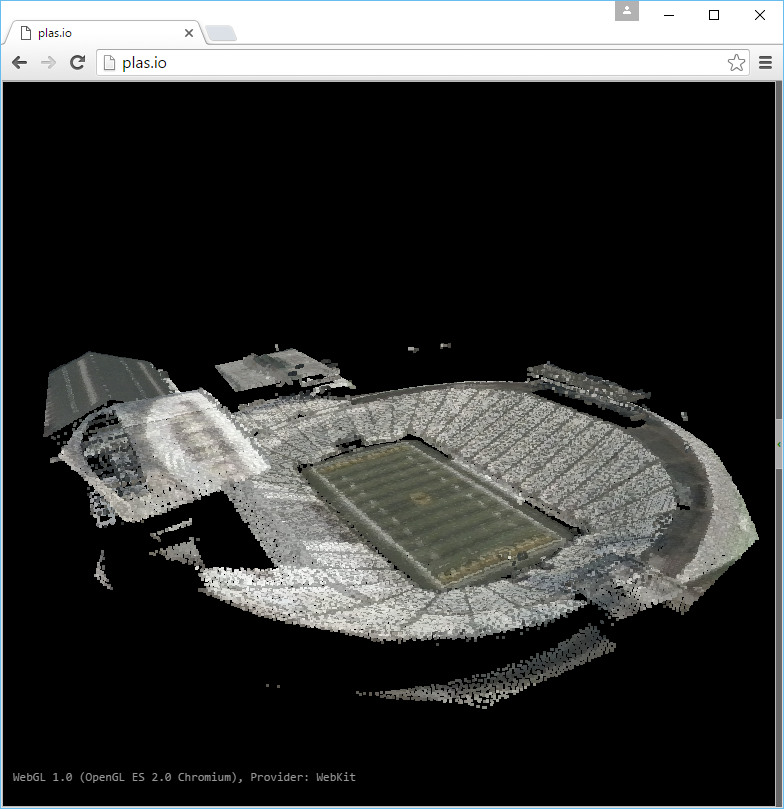
Visualization
Use one of the point cloud visualization tools you installed to take a look at
your ./exercises/analysis/clipping/stadium.las output.
In the example below, we opened the file to view it using the http://plas.io
website.
Notes
filters.overlay does point-in-polygon checks against every point that is read.
Points that are on the boundary are included.